 by Atmaram Yarlagadda, MD; Kevin Swift, PhD; Nabarun Chakraborty, MS, MBA; Rasha Hammamieh, PhD; Amina Abubakar, PharmD; Marianna Wilbur, PharmD; and Anita H. Clayton, MD
by Atmaram Yarlagadda, MD; Kevin Swift, PhD; Nabarun Chakraborty, MS, MBA; Rasha Hammamieh, PhD; Amina Abubakar, PharmD; Marianna Wilbur, PharmD; and Anita H. Clayton, MD
Dr. Yarlagadda is Installation Director of Psychological Health at McDonald Army Health Center in Fort Eustis, Virginia. Drs. Swift, Chakraborty, and Hammamieh are with Medical Readiness Systems Biology, Walter Reed Army Institute of Research in Silver Spring, Maryland. Drs. Abubakar and Wilbur are with Avant Institute in Charlotte, North Carolina. Dr. Clayton is with Department of Psychiatry and Neurobehavioral Sciences, University of Virginia School of Medicine in Charlottesville, Virginia.
Funding: No funding was provided for this article.
Disclosures: Dr. Clayton has received grants from Daré Bioscience; Janssen; Neumora Therapeutics; Otsuka; Relmada Therapeutics, Inc.; and Sage Therapeutics; advisory board fees/consultant fees from AbbVie, Inc.; Biogen; Brii Biosciences; Fabre-Kramer; Initiator Pharma; Janssen Research & Development, LLC; Mycomedica Life Sciences; PureTech Health; Reunion Neuroscience, Inc. (formerly Field Trip Health); S1 Biopharma; Sage Therapeutics; Sertsei Pharmaceuticals, Inc.; and Vella Bioscience, Inc.; royalties/copyright from Ballantine Books/Random House; Changes in Sexual Functioning Questionnaire; and Guilford Publications; and shares/restricted stock units from Euthymics; Mediflix LLC; and S1 Biopharma within the last 12 months. All other authors have no conflicts of interest relevant to the contents of this article.
Innov Clin Neurosci. 2023;20(10–12):12–17.
Abstract
Point-of-care genetic testing for single nucleotide polymorphisms (SNPs) to improve psychiatric treatment in outpatient settings remains a challenge. The presence or absence of certain genomic alleles determines the activity of the encoded enzymes, which ultimately defines the individual’s drug metabolism rate. Classification of poor metabolizers (PMs) and rapid/ultrarapid metabolizers (RMs/UMs) would facilitate personalization and precision of treatment. However, current pharmacogenomic (PGx) testing of multiple genes is comprehensive and requires quantitative analyses for interpretations. We recommend qualitative, fast-track, point-of-care screenings, which are one- or-two gene-based analyses, as a quick initial screening tool to potentially eliminate the need for an expensive quantitative send-out test, which is a costly and lengthy process. We speculate that these tests will be relevant in two major scenarios: 1) clinical psychiatry for treating disease states such as major depressive disorder (MDD) and posttraumatic stress disorder (PTSD), where trial and error is still the mainstay of drug selection and symptom management, a process that is associated with significant delay in optimizing individualized treatment and dose, and thus response; and 2) pain management, where quickly determining an effective level of analgesia while avoiding a toxic level can cause a drastic improvement in mental health.
Keywords: Precision medicine, pharmacogenomics, CYP2D6, CYP2C19, SNP, MDD, PTSD, pain, addictions, suicides
Major depressive disorder (MDD) and posttraumatic stress disorder (PTSD) are psychological conditions commonly resulting from trauma exposure in civilian and military populations.1 Although MDD can present independently of PTSD, MDD and PTSD are commonly comorbid.1 Moreover, epidemiological estimates suggest that the incidence of MDD and PTSD are rising, with 52 percent of diagnosed PTSD cases having comorbid MDD.2 Of note, this percentage is only derived from reported and diagnosed cases. Many individuals are left undiagnosed and/or are afraid to report symptoms due to social stigma and the fear of limiting their social and professional lives. These limiting factors frequently prevent appropriate diagnosis, thus precluding or delaying treatment until symptoms are more severe. Altogether, these factors suggest that PTSD and MDD are a growing concern and present a greater challenge and case load in primary and secondary care.
The available therapeutic options to treat MDD and PTSD are suboptimal in terms of the percentage of patients achieving remission and time to remission. Selective serotonin reuptake inhibitors (SSRIs) are current first-line treatments for MDD and daytime symptoms of PTSD. However, their failure rates approach 40 percent, and as many as 80 percent of patients who experience a positive response fail to achieve full remission.3 Moreover, SSRIs often require trial and error with several different drugs before finding a medication that is effective and well tolerated by the patient. Subsequent dosage titration is often also necessary and can be an ongoing process to optimize therapeutic benefit and encourage patient adherence to the regimen. Each alteration in drug type or dosage can require 3 to 8 weeks between adjustments to reach full therapeutic effect.4 As such, patients that respond to SSRIs often wait weeks to months before experiencing symptom relief, whereas nonresponders go through a similar process but do not experience alleviation of symptoms and must repeat the process. Failure to respond to two or more medication trials of adequate dose and duration is defined as treatment-resistant depression (TRD).5
The lack of a standardized approach that is fully efficacious for all patients may contribute to the growing number of secondary health concerns surrounding MDD and PTSD. Nonadherence due to intolerance, sexual side effects that potentially impact self-esteem, and delays in dose optimization top the list of complications associated with trial and error. Self-medication, overmedication, and substance use disorders are commonly comorbid with MDD and PTSD as patients seek more immediate relief from symptoms.6 Additionally, adverse drug interactions between psychotropic medications are a growing concern, especially among veterans with PTSD. Mohamed and Rosenheck,7 reviewing prescriptions for Veterans’ Administration (VA) patients, noted that in addition to the principal psychotropic medication prescribed to 80 percent of patients, large subsets were given additional drugs, including antidepressants (89%), anxiolytics/sedative hypnotics (61%), and antipsychotics (34%), which are associated with an array of potential side effects and drug-drug or drug-gene interactions. Furthermore, a number of studies attempting to find novel alternatives for SSRIs have ended without success.8 Altogether, these findings underscore the limited chance for success of current options and potential risks of overprescribing psychotropic medications.
Currently, routine genetic testing is rarely utilized in clinical practice. The United States (US) Food and Drug Administration (FDA) has already released resources for physicians outlining genetic variants and their potential gene-drug interaction and likelihood of an adverse event (AE).9 In some cases, these AEs carry significant risks to patient health and safety, and testing is required before a physician can prescribe the drug. Finally, genetic testing is becoming increasingly accessible to patients outside of the clinic.10 Commercial options have popularized affordable genetics for personal screening and genealogical purposes. While it is unclear how data from these companies will integrate with and guide clinical practice, these data have informed numerous scientific studies and increased the understanding of genetic testing.
Discussion
Personalized, or precision, medicine is often considered the ideal healthcare approach and a potential improvement to patient care, especially in patients who do not respond to initial treatments. Certain aspects of clinical practice have benefited from genetic testing. Preventative screening in infants is widely utilized for early detection of trisomy 18 syndrome and Down syndrome.11 Genetic screening in adults can help assess the risk for multifactorial conditions wherein environmental factors and genetic predisposition can both contribute to disease (e.g., BRCA1 and breast cancer12). Carrier testing to identify genes associated with familial disorders, such as cystic fibrosis and sickle cell anemia, has aided in family planning. In addition, more emphasis is being placed on pharmacogenomic (PGx) testing in the clinic. Some branches of medicine, namely oncology, have successfully integrated precision medicine techniques, such as whole genome sequencing, into practice. The field of oncogenomics has improved tumor diagnostics by identifying genomic aberrations driving tumor growth to provide treatment options with increased effectiveness and less toxicity.
Other medical fields, psychiatry included, have been slow to adopt precision medicine techniques into clinical practice, despite physicians displaying largely positive interest.13 That is not to say that psychiatric genomics have not made significant strides in understanding the genetic risk of serious mental health disorders, including MDD and PTSD. Large scale genome-wide association studies (GWAS) have identified allelic variations contributing to the vulnerability to or development of psychiatric conditions. In MDD, several initial GWAS identified candidate single nucleotide polymorphisms (SNPs) that achieved genome-wide significance14,15 but were not replicated by subsequent meta-analyses from multiple sites.16 Later studies, generally with larger samples, identified previously unknown SNPs linked to MDD that were later confirmed by independent data.17–19 Additional studies have evaluated the interaction between multiple SNPs and metabolic pathways to identify gene networks to understand how multiple loci interact to contribute to MDD heritability.20
Large-scale (200,000+ participants) GWAS in PTSD have identified several SNPs associated with PTSD development, which were later replicated by independent data sets.21 Additionally, as PTSD is contingent on a precipitating traumatic event, genetic factors that convey increased risk taking, and thus a putative increased likelihood of trauma, have been identified as risk factors for PTSD.22 Other genetic mediating factors that convey resilience or susceptibility to PTSD have also been identified.23 Despite MDD and PTSD being commonly comorbid, recent large scale GWAS did not identify overlapping genetic risk factors between PTSD and MDD,24,25 although this has been contrasted by twin studies of PTSD and MDD, which have found a strong correlation.26 In both disorders, the genetic component appears to be polygenic, involving numerous loci where variable presence or absence form complex interactions, leading to a wide array of phenotypic manifestations.
Knowledge of the genetic risk factors of psychiatric disorders and opioid abuse has shown usefulness in understanding heritability but has not translated to improved patient care. The pharmacokinetics of a particular drug or the best dose of a drug have significant interindividual and interethnic variability. Pharmacogenomics has examined how the presence or absence of a genomic allele determines the activities of its encoded enzymes, which ultimately defines the individual’s drug metabolism rate. Notably, the cytochrome P450 (CYP) family of enzymes metabolizes about 75 percent of all drugs prescribed today in medical practice and plays an important role in defining selection, optimization of dose, and prevention of toxicity.27 Allelic variations in CYP family subenzymes significantly affect an individual’s overall metabolic profile, which can be categorized into poor, normal, rapid, and ultrarapid metabolizers. These differing metabolic profiles play a substantial role in the trial and error and dose titration of psychotropic drugs, including SSRIs.28 Depending on if a medication is a pro-drug, meaning it must be converted to the active form, or not, a given individual may experience toxicity from a medication or no therapeutic benefit.29 For a pro-drug, such as codeine, rapid metabolizers could experience a buildup of the active form of the medication and possibly toxic effects of the medication, whereas poor metabolizers could experience no therapeutic effect.30 For a drug already in its active form, such as SSRIs, rapid metabolizers could experience no therapeutic effect, while poor metabolizers may experience a buildup of the drug leading to toxicity. For example, individuals with increased function of CYP2C19 may be rapid metabolizers for SSRIs and receive less therapeutic benefit, and as such, are at increased risk of suicide.31–34 On the other hand, individuals with increased function of CYP2D6 may be rapid metabolizers for codeine and experience toxic effects as codeine metabolizes to morphine, whereas those that are poor metabolizers may require a higher dose to achieve therapeutic benefit.20 Therefore, screening for poor and ultrarapid metabolizers is highly recommended during point-of-care analyses before prescribing medications affected by CYP2D6 or CYP2C19. This will provide valuable information that makes it possible to design a more successful drug treatment in such individuals with psychiatric or pain conditions without causing delay or harm.35
Classification
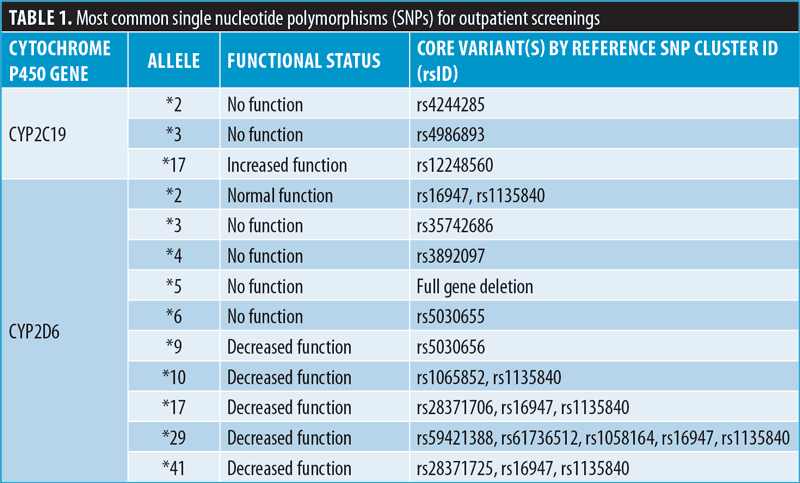
Identifying CYP SNPs that contribute to the metabolic profile of psychotropic drugs, particularly SSRIs, is essential for improving care in patients with MDD and PTSD. The enzymes CYP2D6 and CYP2C19 possess wide allelic variation, which are associated with differing functional status (Table 1). Together CYP2D6 and CYP2C19 metabolize about 60 percent of currently prescribed psychotropic medications.36 Importantly, these enzymes metabolize SSRIs used in treating MDD and PTSD, including citalopram, escitalopram, fluvoxamine, paroxetine, and sertraline.37 Understanding a patient’s metabolic profile by identifying CYP2D6 and CYP2C19 at the point of care could assist the physician in significantly reducing drug trial and error and dose titration.
At the same time, several limitations currently hinder widespread genetic testing in the clinic. In many instances, genetic testing is largely considered an optional or tertiary diagnostic tool, particularly in psychiatry. Thus, genetic testing is often not directly available upon physician request like an MRI or standard blood panel. The limited availability also means that genetic tests are often not covered by insurance providers. The cost of testing is then passed onto patients, who are often reluctant or unable to absorb the cost of an additional test. Also, genetic testing often requires additional time to obtain test results—many clinical settings are not equipped to perform comprehensive genetic screening onsite and must send samples offsite for analysis, which can take one to two weeks. For patients experiencing severe symptoms, this represents a longer period between evaluation and initiation of personalized treatment to achieve symptom relief. Finally, genetic testing requires proper interpretation to inform patient treatment. As such, clinics must have personnel and resources to interpret results, as few medical professionals are equipped to interpret PGx test results. This is mostly due to PGx testing historically not being addressed in the medical education curriculum. However, patients often appreciate a printout of the results with a verbal explanation and are willing to have lab tests to explain their prior experiences and the reasoning behind their individualized treatment plan.
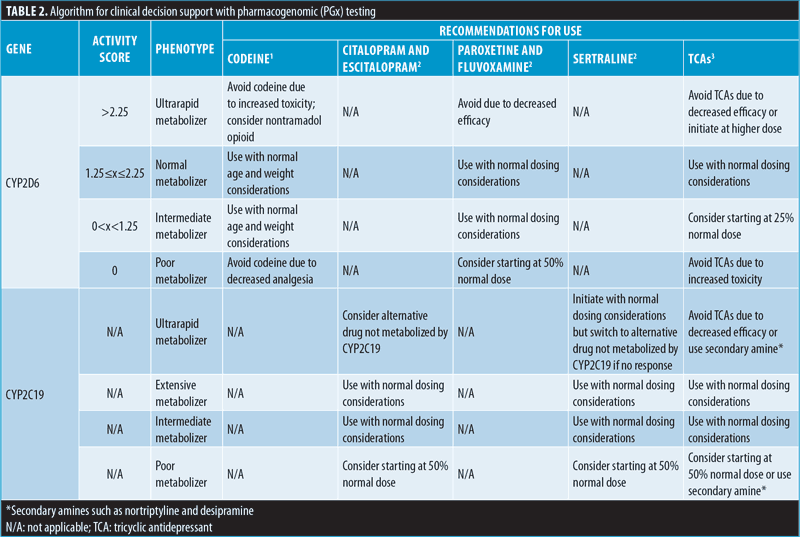
Not only have high-evidence allelic variations for CYP2D6 and CYP2C19 been established, but also guidance is available for how to alter medication regimens based on these allelic variations. The Clinical Pharmacogenetics Implementation Consortium (CPIC) provides this guidance on choosing and dosing medications affected by CYP2D6 and CYP2C19.37–39 For CYP2D6, recommendations are given based on an activity score depending on which two alleles a patient has, which represents the translation of genotype to phenotype.40 Similarly, a systematic nomenclature based on the functional status is used for CYP2C19 and recommendations are given based on a patient’s phenotype.41 There are currently more complicated phenotypes identified for CYP2D6, hence the need for an activity scoring system on which to base medication recommendations (Table 2).
Recommendation
To combat these limitations and reduce the trial and error of drugs and dosage, we propose the development of qualitative fast-track point-of-care tests. These tests are qualitative in nature and designed to determine basic features of CYP2D6 and CYP2C19 activity and any allelic variations that could alter an individual’s metabolic profile. The tests would utilize saliva or a buccal swab to collect patient samples. These swabs present little-to-no risk to patients, require minimal equipment, and can be administered by an allied health professional without requiring a blood draw. Patient samples would then be mixed with necessary test reagents, the dipstick assay placed in the solution, and in 10 to 15 minutes, deliver results determining the presence/absence of allelic variation of CYP enzymes.
Qualitative fast-track point-of-care tests would be delivered in a two-tiered format. The first tier is preemptive and designed to be used when a patient initially arrives at the clinic expressing symptoms. Early identification of poor and rapid metabolizers provides clinicians with key information: 1) identifies patients that may be at increased risk for nonresponse to medication, and therefore at increased risk for suicide or toxicity; 2) informs clinicians of the potential risk of prescribing certain drugs due to incompatibility with the patient’s metabolic profile; and 3) determines what starting dose of a drug may be needed to compensate for a patient’s metabolic profile. These data from the preemptive test would provide clinicians with a starting place for which drugs and doses are best suited to address the patient’s symptoms to maximize treatment efficacy and patient safety. Moreover, it would identify patients who are at inherent increased risk for adverse outcomes and worsening suicidal thoughts and allow more directed monitoring and guidance for their treatment plan.
The second tier is reactive and designed to be used in patients who, after beginning a treatment program, experience negative events, such as dosage toxicity or failure to respond due to being treated with a medication that is incompatible with their metabolic profile. A point-of-care test could identify the metabolic profile of these patients, which may help explain a patient’s negative response to treatment.
Both tiers of qualitative fast-track point-of-care tests will identify CYP2C19 and CYP2D6 variant alleles that already have well-established evidence in the following three categories: 1) changes in activity, either poor or rapid metabolizers that affect drug metabolism, 2) high frequency in the population, and 3) actionable reference materials. The Associates for Molecular Pathology has provided recommendations for which alleles have the highest evidence for both CYP2C19 and CYP2D6, which are summarized in Table 1.42,43 In addition to the previously mentioned alleles, duplications of any allele would result in a heightened functional status to either extreme. Testing for this duplication is especially necessary for CYP2D6 variants. Once a patient’s phenotype has been identified, prescribers can reference a summary of dosing recommendations, such as those shown in Table 2.
Benefits
These tests have several advantages. First, they do not require patient samples to be sent offsite for laboratory processing. This decreases the amount of time necessary to receive results, which could be provided within minutes to hours, helping the patient to avoid having to return for subsequent appointments and allowing the provider to prescribe a treatment compatible with their profile more rapidly. Furthermore, by keeping sample analysis onsite, it increases accessibility to the tests (e.g., in rural primary care/medically underserved regions). Dipstick tests are also cheaper than standard genetic testing, as they do not require additional services to process results. Finally, these tests are simple to interpret, partially due to their limited capacity, compared to broad genetic testing, but also because they often provide a binary positive or negative result, similar to pregnancy tests and COVID-19 tests. This allows for rapid interpretation by a provider who has limited training in pharmacogenomics.
The applicability of these tests also extends beyond SSRI usage in MDD and PTSD. CYP2D6 allelic variations also contribute to differing metabolic phenotypes in opioid metabolism. These tests could help avoid tragic events by locating high-risk individuals vulnerable to overdose or those who do not receive sufficient analgesia from opioid treatment. Screening for allelic variation provides the initial opportunity to define drug selection and dosing to account for metabolic variations.
Limitations
Despite the benefits of these tests, it is unlikely, especially with the current state of supporting scientific evidence, that these tests would widely be adopted as standard of care. More likely, they would be used in patients who fail to achieve symptom alleviation with one or more drugs. Even if these tests become standard of care, patients may be reluctant to have their personal genetic information identified and attached to their medical record. Additionally, the limited results of these tests can only partially explain a patient’s response to a drug. Sex, body weight, ethnicity, drug-drug interactions, and numerous other factors all contribute to an individual’s pharmacokinetics.44 Furthermore, CYPs represent only one family of enzymes responsible for drug metabolism with swaths of additional enzymes, alongside their own unique SNPs, contributing to the overall metabolic profile. While these tests provide initial insight, they are not exhaustive and will not fully explain a patient’s response to a drug.
Conclusion
Despite limitations, point-of-care CYP testing has significant potential to improve patient care in psychiatry and beyond as personalized medicine advances. Current research in disease biomarkers is poised to allow unparalleled diagnostic capabilities to assess risks for disease development and progression. Additionally, therapy that targets and corrects genetic aberrations for a myriad of conditions is becoming commercially available. While there is debate as to whether these advances will improve psychiatric treatment, the current treatment options—particularly for MDD and PTSD—are not fully effective. The proposed point-of-care tests would enable providers to better personalize the current treatment options to address patient symptoms by decreasing drug and dose trial and error. Furthermore, the information from these tests may improve patient adherence and prevent attrition by providing the patient with personalized data informing their treatment, rather than trial and error.
Acknowledgments
We would like to thank Stephen Furry for administrative support for the manuscript.
Disclaimer
The material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and publication. The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or reflecting the view of the Department of the Army or the Department of Defense.
References
- Flory JD, Yehuda R. Comorbidity between post-traumatic stress disorder and major depressive disorder: alternative explanations and treatment considerations. Dialogues Clin Neurosci. 2015;17(2):141–150.
- Rytwinski NK, Scur MD, Feeny NC, Youngstrom EA. The co-occurrence of major depressive disorder among individuals with posttraumatic stress disorder: a meta-analysis. J Trauma Stress. 2013;26(3):299–309.
- Berger W, Mendlowicz MV, Marques-Portella C, et al. Pharmacologic alternatives to antidepressants in posttraumatic stress disorder: a systematic review. Prog Neuropsychopharmacolo Biol Psychiatry. 2009;33(2):169–180.
- Leuchter AF, Cook IA, Hunter AM, Korb AS. A new paradigm for the prediction of antidepressant treatment response. Dialogues Clin Neurosci. 2009;11(4):435–446.
- Souery D, Papakostas GI, Trivedi MH. Treatment-resistant depression. J Clin Psychiatry. 2006;67(6):16–22.
- Jacobsen LK, Southwick SM, Kosten TR. Substance use disorders in patients with posttraumatic stress disorder: a review of the literature. Am J Psychiatry. 2001;158:1184–1190.
- Mohamed S, Rosenheck RA. Pharmacotherapy of PTSD in the US Department of Veterans Affairs: diagnostic- and symptom-guided drug selection. J Clin Psychiatry. 2008;69(6):959–965.
- Asnis GM, Kohn SR, Henderson M, Brown NL. SSRIs versus non-SSRIs in post-traumatic stress disorder: an update with recommendations. Drugs. 2004;64(4):383–404.
- United States Food and Drug Administration. Table of pharmacogenetic associations. Current as of 26 Oct 2022. https://www.fda.gov/medical-devices/precision-medicine/table-pharmacogenetic-associations. Accessed 30 Jan 2023.
- Annas GJ, Elias S. 23andMe and the FDA. N Engl J Med. 2014;370(11):985–988.
- Loane M, Morris JK, Addor MC, et al. Twenty-year trends in the prevalence of Down syndrome and other trisomies in Europe : impact of maternal age and prenatal screening. Eur J Hum Genet. 2013;21(1):27–33.
- Fackenthal JD, Olopade OI. Breast cancer risk associated with BRCA1 and BRCA2 in diverse populations. Nat Rev Cancer. 2007;7(12):937–948.
- Unertl KM, Jaffa H, Field JR, et al. Clinician perspectives on using pharmacogenomics in clinical practice. Per Med. 2015;12(4):339–347.
- Kohli MA, Lucae S, Saemann PG, et al. The neuronal transporter gene SLC6A15 confers risk to major depression. Neuron. 2011;70(2):252–265.
- Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium; Ripke S, Wray NR, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18(4):497–511.
- Shadrina M, Bondarenko EA, Slominsky PA. Genetics factors in major depression disease. Front Psychiatry. 2018;9:334.
- Consortium C. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523(7562):588–591.
- Okbay A, Baselmans BML, De Neve JE, et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet. 2016;48(6):624–633.
- Wray NR, Ripke S, Mattheisen M, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50(5):668–681.
- Song GG, Kim JH, Lee YH. Genome-wide pathway analysis in major depressive disorder. J Mol Neurosci. 2013;51(2):428–436.
- Stein MB, Levey DF, Cheng Z, et al. Genome-wide association analyses of post-traumatic stress disorder and its symptom subdomains in the Million Veteran Program. Nat Genet. 2021;53(2):174–184.
- Polimanti R, Ratanatharathorn A, Maihofer AX, et al. Association of economic status and educational attainment with posttraumatic stress disorder a mendelian randomization study. JAMA Netw Open. 2019;2(5):1–17.
- Nievergelt CM, Maihofer AX, Klengel T, et al. International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nat Commun. 2019;10(1):1–16.
- Duncan LE, Ratanatharathorn A, Aiello AE, et al. Largest GWAS of PTSD (N=20070) yields genetic overlap with schizophrenia and sex differences in heritability. Mol Psychiatry. 2018;23(3):666–673.
- Anttila V, Bulik-Sullivan B, Finucane HK, et al. Analysis of shared heritability in common disorders of the brain. Science. 2018;360(6395): eaap8757.
- Sartor CE, Grant JD, Lynskey MT, et al. Common heritable contributions to low-risk trauma, high-risk trauma, posttraumatic stress disorder, and major depression. Arch Gen Psychiatry. 2012;69(3):293–299.
- Guengerich FP. Cytochrome P450 and chemical toxicology. Chem Res Toxicol. 2008;21(1):70–83.
- Probst-Schendzielorz K, Viviani R, Stingl JC. Effect of Cytochrome P450 polymorphism on the action and metabolism of selective serotonin reuptake inhibitors. Expert Opin Drug Metab Toxicol. 2015;11(8):1219–1232.
- Begg EJ, Helsby NA, Jensen BP. Pharmacogenetics of drug-metabolizing enzymes: the prodrug hypothesis. Pharmacogenomics. 2012;13(1):83–89.
- Dean L, Kane M. Codeine therapy and CYP2D6 genotype. In: Pratt VM, Scott SA, Piromhamed M, et al, eds. Medical Genetics Summaries [internet]. National Center for Biotechnology Information (US); 2021:213–230.
- Peñas-Lledó EM, Blasco-Fontecilla H, Dorado P, et al. CYP2D6 and the severity of suicide attempts. Pharmacogenomics. 2012;13(2):179–184.
- Peñas-Lledó EM, Dorado P, Agüera Z, et al. High risk of lifetime history of suicide attempts among CYP2D6 ultrarapid metabolizers with eating disorders. Mol Psychiatry. 2011;16(7):691–692.
- Peñas-Lledó EM, Guillaume S, Naranjo MEG, et al. A combined high CYP2D6-CYP2C19 metabolic capacity is associated with the severity of suicide attempt as measured by objective circumstances. Pharmacogenomics J. 2015;15(2):172–176.
- Kawanishi C, Lundgren S, Agren H, Bertilsson L. Increased incidence of CYP2D6 gene duplication in patients with persistent mood disorders: ultrarapid metabolism of antidepressants as a cause of nonresponse. A pilot study. Eur J Clin Pharmacol. 2004;59(11):803–807.
- Ingelman-Sundberg M, Oscarson M, McLellan RA. Polymorphic human cytochrome P450 enzymes: an opportunity for individualized drug treatment. Trends Pharmacol Sci. 1999;20(8):342–349.
- Abubakar A, Bentley O. Precision medicine and pharmacogenomics in community and primary care settings. Pharm Today. 2018;24(2):55–68.
- Hicks JK, Sangkuhl K, Swen JJ, et al. Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther. 2017;102(1):37–44.
- Hicks JK, Bishop JR, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther. 2015;98(2):127-134. doi:10.1002/CPT.147
- Crews KR, Monte AA, Huddart R, et al. Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6, OPRM1, and COMT genotypes and select opioid therapy. Clin Pharmacol Ther. 2021;110(4):888–896.
- Caudle KE, Sangkuhl K, Whirl-Carrillo M, et al. Standardizing CYP2D6 genotype to phenotype translation: consensus recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin Transl Sci. 2020;13(1):116–124.
- Botton MR, Whirl-Carrillo M, Del Tredici AL, et al. PharmVar GeneFocus: CYP2C19. Clin Pharmacol Ther. 2021;109(2):352–366.
- Pratt VM, Del Tredici AL, Hachad H, et al. Recommendations for clinical CYP2C19 genotyping allele selection: a report of the Association for Molecular Pathology. J Mol Diagnostics. 2018;20(3):269–276.
- Pratt VM, Cavallari LH, Del Tredici AL, et al. Recommendations for clinical CYP2D6 genotyping allele selection: a joint consensus recommendation of the Association for Molecular Pathology, College of American Pathologists, Dutch Pharmacogenetics Working Group of the Royal Dutch Pharmacists Association, and the European Society for Pharmacogenomics and Personalized Therapy. J Mol Diagnostics. 2021;23(9):1047–1064.
- Burger D, van der Heiden I, Porte C, et al. Interpatient variability in the pharmacokinetics of the HIV non-nucleoside reverse transcriptase inhibitor efavirenz: the effect of gender, race, and CYP2B6 polymorphism. Br J Clin Pharmacol. 2005;61(2):148–154.


