by Erica A. Smith, PhD; William P. Horan, PhD; Dominique Demolle, PhD; Peter Schueler, MD; Dong-Jing Fu, MD, PhD; Ariana E. Anderson, PhD; Joseph Geraci, MSc, PhD; Florence Butlen-Ducuing, MD, PhD; Jasmine Link, MS; Ni A. Khin, MD; Robert Morlock, PhD; and Larry D. Alphs, MD, PhD
Drs. Smith and Demolle are with Cognivia in Mont St. Guibert, Belgium. Dr. Horan is with VeraSci in Durham, North Carolina. Drs. Horan and Anderson are with the University of California at Los Angeles in Los Angeles, California. Dr. Schueler is with ICON Clinical Research in Langen, Germany. Dr. Fu is with Janssen Research and Development, LLC, in Titusville, New Jersey. Dr. Geraci is with Nurosene Health, Inc. in Toronto, Ontario; Queen’s University in Kingston, Ontario; and the Center for Biotechnology and Genomics Medicine, Medical College of Georgia, in Augusta, Georgia. Dr. Butlen-Ducuing is with European Medicines Agency in Amsterdam, Netherlands. Ms. Link is with Boehringer Ingelheim Pharma GmbH and Company KG in Baden Württenberg, Germany. Dr. Khin is with Neurocrine Biosciences, Inc. in San Diego, California. Dr. Morlock is with YourCareChoice in Ann Arbor, Michigan. Dr. Alphs is with Denovo Biopharma, LLC in San Diego, California (at the time of writing, he was with Newron Pharmaceuticals in Morriston, New Jersey).
Funding: No funding was provided for this study.
Disclosures: Drs. Smith and Demolle are employees of Cognivia, which develops commercial placebo prediction products. Dr. Horan is a full-time employee of VeraSci. Dr. Fu is an employee of Janssen Research and Development, LLC, and is a stockholder of Johnson & Johnson, Inc. Dr. Geraci is a significant shareholder of NetraMark Corp. and is an employee of Nurosene Health, Inc., which develops commercial placebo prediction products. Ms. Link is an employee of Boehringer Ingelheim International GmbH but did not receive any direct compensation relating to the development of this manuscript. Dr. Alphs is an employee of and has stock in Newron Pharmaceuticals and has stock in Johnson & Johnson, Inc.
Innov Clin Neurosci. 2022;19(1–3):60–70.
ABSTRACT: The placebo response is a highly complex psychosocial-biological phenomenon that has challenged drug development for decades, particularly in neurological and psychiatric disease. While decades of research have aimed to understand clinical trial factors that contribute to the placebo response, a comprehensive solution to manage the placebo response in drug development has yet to emerge. Advanced data analytic techniques, such as artificial intelligence (AI), might be needed to take the next leap forward in mitigating the negative consequences of high placebo-response rates. The objective of this review was to explore the use of techniques such as AI and the sub-discipline of machine learning (ML) to address placebo response in practical ways that can positively impact drug development. This examination focused on the critical factors that should be considered in applying AI and ML to the placebo response issue, examples of how these techniques can be used, and the regulatory considerations for integrating these approaches into clinical trials.
Keywords: placebo response, placebo effect, clinical trials, artificial intelligence, machine learning, machine intelligence
Double-blind, placebo-controlled, randomized clinical trials (RCTs) are the gold standard for the evaluation of experimental therapeutics, but comparisons of efficacy and safety to those of placebo treatment pose unique and specific challenges. The placebo response, the phenomenon by which clinical trial patients experience a clinical improvement after treatment with a placebo, has historically made demonstration of efficacy difficult, leading to increased data variability, clinical trial failures, and potential abandonment of compounds that may otherwise be efficacious. For example, meta-analyses of depression studies have shown that high placebo response, not low medication response, is responsible for most of the change in drug-placebo differences over time.1,2 High placebo-response rates are seen across therapeutic areas but are particularly high in neurological and psychiatric diseases, and substantial attention has been given to placebo response in areas such as pain, depression, schizophrenia, and Parkinson’s disease. This is confounded by the use of primarily subjective, patient-reported outcomes as primary endpoints for these indications.
Sponsors of clinical trials have employed a variety of methods to control or otherwise limit the impact of the placebo response, focused on identifying high placebo responders and excluding them from the trial; minimizing inflation of patient expectation, either by clinical trial design factors or site/staff training; or reducing variation or errors in either patient or physician/rater reporting of outcomes. While these strategies have yielded some benefit to trials, the problem persists, with some reports suggesting that the placebo response is increasing over time in areas such as pain3 and psychiatry.4–7 Clearly, there is still a substantial need for new approaches to address the placebo response in drug development, and it is critical for future approaches to be scalable for large, industrial clinical trials and acceptable to regulatory agencies.
These efforts to understand and reduce the impact of the placebo response in drug development have been complicated by the complexity of the placebo response itself. The wealth of basic scientific research performed in this area has illuminated the wide variety of factors that contribute to the placebo response, ranging from biological factors (e.g. activation of specific circuitry in the brain) to social cues (e.g. transfer of expectation from investigators to patients) and beyond. The placebo response is dynamic, changes over time, varies in different cultures and geographies, and can be influenced by the clinical trial itself. Further adding to this complexity, while the placebo response is primarily measured in placebo-treated patients, it also constitutes some component of the treatment effect in drug-treated patients. This contribution has been estimated to comprise about 65 percent of the measured treatment effect in pain8 and depression9 trials.
Considering the complexity and dynamic nature of the placebo response, the advent of advanced and sophisticated data analytic approaches could be helpful, or even required, to shed new light on this old problem. Advances in artificial intelligence (AI), specifically machine learning (ML)-based approaches, have made these techniques more accessible and acceptable, and their adoption by the biopharmaceutical industry is apparent in areas such as target validation, computational chemistry, and drug repurposing. The utilization of methods such as ensemble trees (e.g., boosted trees and random forest), support vector machines, and deep neural networks are among the most popular methods being used. While there are many specific methods available, the essential character of ML algorithms is that, in some sense, they program themselves. This means that the algorithms learn about some set of data and are not manually programmed to do a task, but instead, models emerge from their interaction with data. Here, we explore the role and potential of AI in understanding and even predicting the placebo response.
With this in mind, the goals of this review are as follows:
- Inform scientists and physicians conducting clinical trials about AI.
- Explore how AI could be used to address placebo response in innovative, productive, and practical ways.
- Examine how AI has been or is currently being used to understand or address the challenges of the placebo response in neuroscience.
- List the key regulatory questions that emerge for the AI approaches that could be, have been, and are being used to address the placebo response challenge in regulated trials.
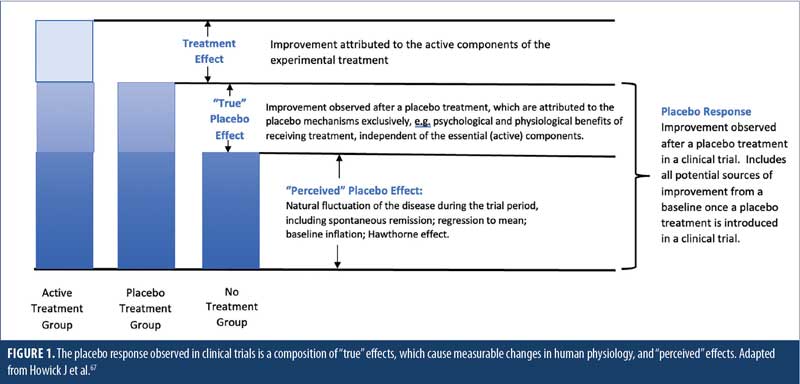
It is important to lay out our consensus definitions for the terms used throughout this manuscript. Terms are defined in Table 1, with Figure 1 illustrating the differences between placebo response, real and perceived placebo effect, and treatment effect. It is noteworthy that, for the sake of this review, the term “placebo response” will be used to indicate the full range of responses that a patient receiving a placebo might exhibit, from negative (nocebo) response to no response to positive (placebo) response. This is important, as AI-based analytical or predictive approaches should aim to model this entire spectrum of response. Also, the term “AI” will be used to refer to the entire scope of machine intelligence/artificial intelligence, including ML. In areas where ML is specified, it is referring to only the sub-discipline of ML and not the full field of AI.

Factors Contributing to the Placebo Response
A wide range of factors can contribute to the apparent benefit in a specific endpoint assessed in trial participants who receive an inactive treatment (Figure 2). These can be broadly categorized into treatment-related, natural history, and measurement factors.10 Treatment-related, or real, factors, including intrinsic psychological experiences of participants and extrinsic psychosocial features of the trial environment, are fundamental contributors.11,12 Chief among the intrinsic factors are participant expectancies, such as how much they believe they will benefit as a consequence of a treatment.13 Intrinsic factors also include the relevant learning histories of participants, such as prior treatment successes or failures, and even conditioned associations between symptom improvements and salient treatment-related cues, such as pills and medical procedures. These intrinsic psychological processes interact with various extrinsic trial features to impact the magnitude of placebo response. For example, placebo response could be larger in trials in later stages of development or with more frequent and/or longer study visits. Furthermore, frequent contact and attention provided by empathetic professionals at trial sites can foster an interpersonal environment that promotes nonspecific treatment benefits. The magnitude of placebo response also relates to information provided to participants on trial design, such as the number of treatment arms (i.e., probability of receiving placebo), and even to physical characteristics of the treatment itself (e.g., mode of administration, the color or size of pills).14 The invasiveness and cost of treatment create higher expectations for efficacy and thus higher placebo response. Notably, the impact of all these treatment-related factors varies among individuals and is likely moderated by certain participant characteristics.15,16 Individual differences in variables, ranging across demographics, disease characteristics, personality traits, genetics, and culture, have been investigated in this context.
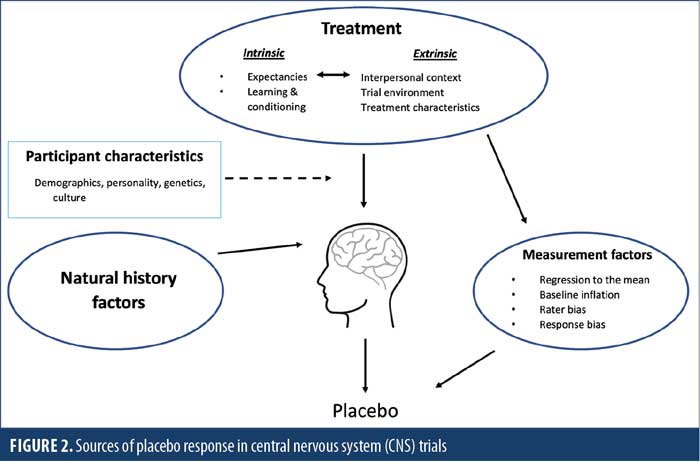
Several other factors beyond these treatment-related factors can also contribute to the measured placebo response over the length of a clinical trial, including natural history factors, which refer to the progression of the disease over time in the absence of treatment. For example, participants often enter trials when their symptoms are relatively severe and might then experience decreases or spontaneous fluctuations in symptoms over time that simply reflect the natural course of illness. Furthermore, central nervous system (CNS) trial endpoints often rely on subjective ratings, which are prone to various sources of error related to measurement factors. One key source is the statistical phenomenon of regression to the mean, or the tendency for individual outcomes with more extreme initial scores to shift toward the group mean over repeated measurements. Biases in how raters and participants approach the assessment also impact measurement. For example, raters can be biased by their own beliefs about an experimental treatment or by other issues, such as financial incentives to enroll patients, which can lead to inflated baseline symptom ratings. Alternatively, well-known response biases, such as providing responses that are socially desirable or aligned with beliefs about what the expert investigators hope to find, can influence how participants report their symptoms.
Our current understanding of the placebo response comes largely from isolated clinical studies, in which one or a small number of factors are parametrically altered to understand their influence on patient response to a placebo, or from meta-analyses of multiple clinical studies in a specific indication. While the field has gained valuable insight with these approaches, it is difficult-to-impossible to obtain either qualitative or quantitative understanding of the complex interplay between the multiple factors that can define or influence the placebo response. We simply do not yet understand how to weigh these factors, how they are interrelated, and what additional factors might impact these relationships. The advent of advanced data analytic techniques, such as AI, along with the predictive modeling approaches made possible by ML, might not only be useful but necessary to begin to understand a phenomenon as complex as the placebo response.
How AI/ML Can Be Used to Understand or Address the Placebo Response in Drug Development
The use of AI (including the subdiscipline of ML) can provide several advantages in addressing placebo response, including the ability to analyze large volumes of data. There are different types of AI that can be useful, ranging from support vector machines (SVM) to artificial neural networks, each with different advantages and limitations. For example, some AI-driven approaches require data from thousands to tens of thousands of patients to draw meaningful conclusions; such applications might be limited to large-scale meta-analyses of multiple clinical trials. Selection of specific AI approaches should ideally be matched to the scientific or research question being pursued.
While AI-based approaches are well-positioned to provide insight and at least partial solutions to address the issue of the placebo response in drug development, the limitations and risks associated with these approaches need to be kept in mind. The choice of specific AI/ML methods can lead to biases in outcome. For example, methods should be matched to the size and characteristics of the dataset to avoid the risk of overfitting—or producing a model that is too closely aligned to a specific dataset and loses applicability to other sets of data. As AI-based algorithms might make unsupervised associations, data should be interpreted by experts in clinical trial design and execution to ensure that relevant conclusions are drawn. Beyond this, all algorithms should be evaluated and conclusions confirmed on independent datasets from the data used for the initial analysis.
One potential application of AI to better understand the placebo response and instruct the field on how to optimize clinical trial design would be to identify key trial design factors associated with high placebo response. This type of approach could be applied retrospectively to large or complex datasets and would be useful in identifying associations that were too subtle or not readily apparent by other methods (e.g., classical statistics). Retrospective meta-analyses could be used to compare trial design features and understand their specific impact on assay sensitivity. For example, it is well-recognized that specific clinical trial design factors, such as the number of treatment arms or trial duration, can increase patient expectancy and thus increase placebo response. AI-based methods could not only identify additional factors that might not be obvious, but could also begin to quantify the relative importance of each factor across trials or between disease areas and their complex dependencies.
Beyond identifying meaningful associations between clinical trial design factors and placebo response, predictive algorithms based on AI that could identify placebo responders at baseline are appealing possibilities. In developing such algorithms using AI, there are a few critical components. First, it is important to select the most pertinent baseline features (perhaps using techniques like SVM recursive feature elimination [SVM-RFE] or data-driven methodologies), noting that some of these features might be disease specific. Particular attention should be paid to limiting the risk of overfitting. To minimize this risk, modeling approaches that are appropriate for the dataset’s characteristics, including sample size, number of features, nonlinearities, and noise distribution, should be selected. Performance of the predictive model should then be appropriately evaluated to confirm modeling choices, with care taken to avoid any bias in the analysis. Common methods to estimate performance on an initial model are cross-validation or repeated random subsampling, in which a portion of the dataset (e.g., 90%) is used to train the model, which is then evaluated on the remaining dataset (e.g., 10%). This process is iterated dozens or hundreds of times to confirm the performance of the model. To further confirm the validity of the model, it could be fixed a priori and evaluated on an independent set of patients other than those used to train the model.
If techniques like AI can eventually be used to predict placebo responders before the first administration of a drug, the next obvious question is, how should that information be used for population enrichment or targeted recruitment? While there are several options, one approach would be to exclude high placebo responders to improve clinical trial assay sensitivity or the degree to which the trial can distinguish the response to treatment from the response to placebo. The best way to estimate the potential impact of this approach is to evaluate the effect of placebo run-in periods. Placebo run-in trial designs, ranging in complexity from single-blind placebo run-in to sequential parallel comparison design (SPCD),17 intend to identify and exclude patients that show a strong improvement with placebo. However, meta-analyses of pain and psychiatry RCTs have demonstrated that placebo lead-in phases neither decreased the placebo response18 nor increased the difference in responses between active drug and placebo groups.19–21 Beyond this, high placebo responders also often have a robust response to drugs, so excluding high placebo responders could actually result in an enriched population of nocebo responders. Considering that, when used as a patient selection factor, placebo run-in designs result in disqualification of 7 to 25 percent of patients,18,19 this approach could extend trial recruiting time and cost for little or no gain.
Excluding placebo responders effectively results in limiting the population in which drug efficacy and safety are evaluated. Thus, extrapolation to a broader population later can lead to regulatory concerns. In fact, use of an SPCD was cited as a potential reason for rejection of a recent application of a drug to treat depression.22 It is possible, however, that using AI or ML to identify placebo responders might be more effective or more efficient than placebo run-in designs, and thus have a more positive impact. Despite these potential drawbacks, there might be situations in which excluding placebo responders could be appropriate (e.g., in early phase studies or as part of fail-fast development strategies).
In exploring how AI can be used to address placebo response in drug development, ethical considerations based on the understanding that placebo response describes clinical improvement in trial participants must be kept in mind. For example, if specific clinical study design characteristics cause clinical improvement in clinical trial subjects, is it ethical to modify study design and limit the benefit to subjects? If a study proposes to remove high placebo responders, is it ethical to restrict access of these patients to experimental therapies? These questions must be balanced against the acknowledgment that the placebo response problem can contribute to high failure rates of clinical trials and drug development programs, and thus negatively impact the availability of new medicines to entire patient populations.
AI-based predictions of placebo responders can potentially be used in other ways that could positively impact clinical trials. One option would be stratifying patients based on their placebo responsiveness. This would address the critical risk of unbalanced distribution of strong placebo responders within the respective treatment arms, especially in clinical trials with small sample sizes. A second option would be the use of predicted placebo responsiveness as a baseline covariate in the statistical analysis. In this way, some of the variation of the outcomes between patients can be explained by this covariate, thus leading to an increase in study power and a reduced risk of clinical trial failure.
Published Examples Using AI to Understand or Predict Placebo Response
A handful of initial published studies have applied AI-based approaches to CNS clinical trial data to understand determinants of placebo response. Three particularly relevant studies come from secondary analyses of antidepressant trials in patients with major depressive disorder (MDD) from three different age groups. All three attempted to identify baseline characteristics that predict placebo response, though the methods and analytic approaches differed considerably.
Using data from an RCT of citalopram versus placebo in 174 older adults with MDD, Zilcha-Mano et al23 applied a random forest ML approach to 11 baseline demographic, clinical, and cognitive variables to identify the strongest overall moderators of response on clinical ratings of depression. The predictor profile differed between conditions: the strongest response (i.e., symptom decrease) in the placebo arm was for patients with higher education (>12 years of school), whereas the strongest response in the drug arm was for patients with lower education (<12 years of school) with a longer duration of illness.
Using an alternative strategy with data from 141 adults with MDD in the placebo arm of the EMBARC trial, Trivedi et al24 applied an elastic net regularization approach to 283 baseline clinical, behavioral, imaging, and electrophysiological variables to identify the most robust parsimonious set of predictors of clinician-rated outcome. Lower baseline depression severity, younger age, absence of melancholic features or history of physical abuse, less anxious arousal, less anhedonia, less neuroticism, and higher average theta current density in the rostral anterior cingulate were retained as the best predictors of placebo response. Focusing on these variables, a Bayesian method was used to develop an interactive calculator to predict the likelihood of placebo response at the individual subject level, which demonstrated a relatively high degree of predictive accuracy within this sample.
Lorenzo-Luaces et al25 applied a similar elastic net regularization approach to 18 baseline demographic, clinical, and cognitive variables in the placebo arm (n=112) of an RCT in adolescent MDD. Baseline depression severity, age, action stage of change, sleep problems, expectations, and maternal depression were retained as key predictors of post-treatment self-reported depression. These variables were used to develop a subject-level prognostic index, reflecting vulnerability to placebo response.
A few other studies have started to use ML approaches to examine issues related to placebo response. For example, ML methods have been used to examine predictors of self-reported treatment expectancies and sudden treatment gains during RCTs for depression.26,27 Using functional magnetic resonance imaging (fMRI) data, exploratory studies (number of subjects≤50) have found links between certain resting state functional connectivity characteristics and placebo response in studies of depression28 and acupuncture for pain.29 In summary, while the application of ML methods to understand placebo response is in its infancy, promising findings, primarily in the area of depression, are starting to emerge.
Emerging Research Using AI to Understand or Predict Placebo Response
As experts in the field, several working group members contributed examples from their own research that, considering the relative newness of this field, have yet to be published in their entireties. These emerging examples are included to share some insight into what might be developing in this area over the next several years. The inclusion of these examples does not indicate preference, only the availability of data for consideration by this team.
Development of a placebo-quantified response score in schizophrenia. Efforts to overcome the adverse impact of high placebo response rates have led to multistage trial designs in which placebo responders are identified after placebo administration. This study hypothesized that 1) a patient’s expected placebo response (placebo quantified response score [PQRS]) could be determined at baseline using models trained on placebo-treated patients within other studies; 2) the PQRS would predict medication response in new studies; and 3) trial enrichment based on PQRS would enable sponsors to increase effect size or reduce the study sample size necessary to achieve a specific effect.
Using only baseline information (age, diagnosis age, sex, country, baseline PANSS scores), random forests ML models were trained and tested across 11 studies (N=3,647) in schizophrenia to predict the placebo response (i.e., to create a PQRS) using placebo-treated patients.30 The medication response was also predicted across studies using baseline information and the PQRS to test whether placebo risk influenced medication response. Several different trial enrichment designs were evaluated using placebo responder exclusion or medication responder inclusion predictions. Omega-squared, eta-squared, and partial eta-squared effect size measures were compared for different screening strategies.
The PQRS predicted the actual placebo response and the medication response (p<0.05) above and beyond baseline information. The average correlation between the PQRS and the actual placebo response across all nine placebo-controlled studies was r=0.24, corresponding to an R2 of 5.9 percent (p<0.001). In the parametric models, a greater PQRS was associated with a larger treatment response within the medication group. The correlation between the PQRS and the total treatment response was r=0.22, corresponding to an R2 of 4.8 percent (p<0.05.)30 PQRS prediction was, however, poor in comparator studies or when study populations had dissimilar demographics.
Associating placebo response with attitudinal variables in bipolar disorder. An application of a machine intelligence method combined with data from a clinical trial (NCT01467700) evaluating an investigational drug for the treatment of acute depressive episode in bipolar 1 disorder demonstrates the potential for AI to assess the impact of placebo response within clinical trials. The analysis focused on 378 patients that completed the study. The study drug did not show separation from placebo, and the team used ML for the prediction of placebo response and nonresponse. The variables utilized in these models were derived from a collection of clinical scales at baseline, including the Montgomery–Asberg Depression Rating Scale (MADRS), Hamilton Anxiety Rating (HAM-A), Young Mania Rating Scale (YMRS), and the Clinical Trial and Site Scale Modified (CTSS-M), a patient-reported assessment designed to assess the probability of placebo response, which were administered in this clinical trial.31
The machine intelligence method deployed was a novel blend of ensemble trees and unsupervised learning to identify responders, determined as patients who had 50-percent improvement in MADRS from baseline. Once clusters were defined, ensemble trees were used to learn from these partitions and to focus on subpopulations where they could make the strongest predictions.32 The training data consisted of one cohort of 64 participants receiving placebo out of 71 who had CTSS-M data at baseline (total placebo participants in the study, N=115).33 Models were tested on 218 out of 239 participants who had baseline CTSS-M and were given the active study drug, as there was no significant separation from placebo. It is noteworthy that the models employed for this work are designed to look for coherent causal subpopulations within complex patient populations, meaning that they made predictions for a few patients. Thus, if the model claimed that a person was a nonresponder, it was correct 87 percent of the time. There were 44 participants for whom it made a nonresponder call. The model being reported here was trained to either make a “No Call” or to identify nonresponders. Specifically, it made a false nonresponder call on five out of 54 responders and made a true nonresponder call on 39 out of 164 patients. Thus, when the model did make a call, it was correct approximately 87 percent of the time, but it only made calls on approximately one-fifth of the patients.33
While the results of this study are promising, they do have some limitations. First, the study considered only participants with bipolar 1 disorder who were having an acute depressive episode, and analysis was limited to patients completing CTSS-M (English speaking United States [US] participants only). Importantly, the models built by this approach do not make predictions for everyone, just for individuals about which the models are confident. As both training and testing of the model were conducted in the same study, these findings require replication in an independent study before definitive conclusions can be drawn.
Prediction of patient placebo responsiveness based on patient psychological traits. The contribution of patient psychology to placebo response has been well-documented in the literature34–42 and might be necessary to robustly predict placebo response. One AI method considers psychological features of the individual patient, along with demographics, disease intensity, and other standard baseline data, and utilizes an ML-based predictive algorithm to predict the placebo response of an individual at baseline. The predictive algorithm used in this approach included ridge regression and linear support vector regression, which are both linear methods. In general, the initial weights and features included in each disease-specific model are first trained on data from placebo-treated patients, and model performance is estimated using repeated random sub-sampling techniques (e.g., Monte Carlo Cross-Validation) and/or evaluated in an independent clinical trial to confirm and validate predictive performance. The predictive model can then be prespecified and used to calculate a score in the clinical study that describes the placebo responsiveness of the patients. This score can be considered a baseline covariate following the regulatory guidances43,44 that, when included in the statistical analysis (ANCOVA), might reduce data variance and increase study power.
Predictive models have been developed in chronic pain, using data from 87 patients with peripheral neuropathic pain (PNP) or a mixed population of 135 patients with PNP or osteoarthritis (OA) pain. These two models exhibited similar performances. Furthermore, the model trained on patients with PNP was only used to predict placebo response in the mixed population of patients with PNP or OA, suggesting that factors predictive of placebo response in patients with PNP and OA are consistent.45
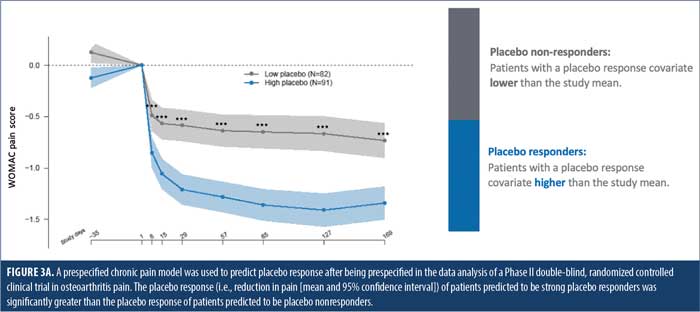
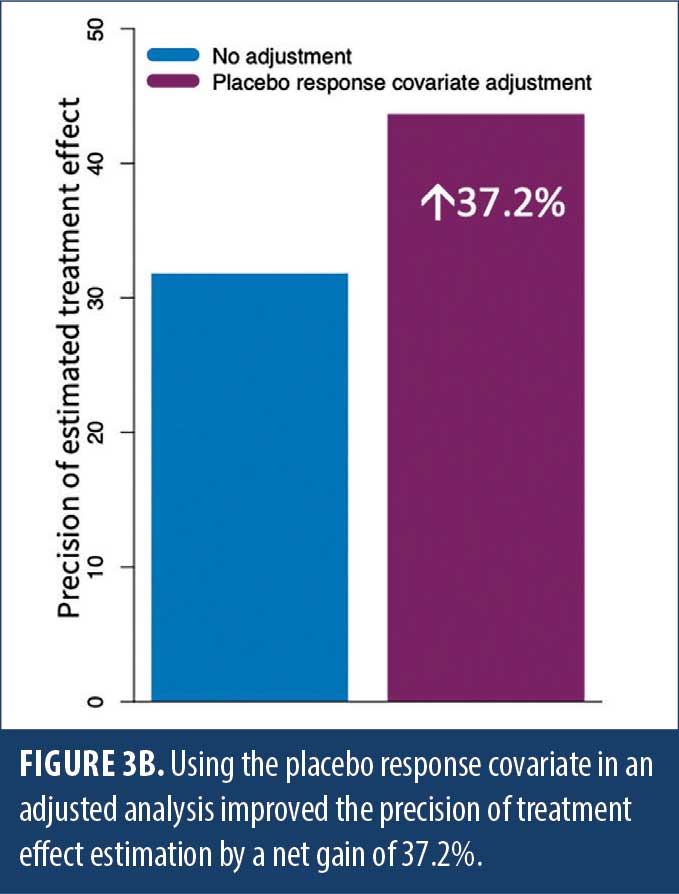
The chronic pain model was trained on data from 211 patients with PNP or OA from several pooled clinical studies. This model was fully prespecified in the statistical analysis plan in a recent Phase II, randomized, placebo-controlled, multicenter trial in OA pain (NCT04129944). In this study, the model predicted placebo response for multiple endpoints, with a Pearson’s correlation ranging from 55.2 to 59.7 percent for the three components of the Western Ontario and McMaster Universities Arthritis Index (WOMAC) battery (R2: 30.4–35.6%, p<0.001).46 The model was further predictive for all patients in the trial (R2: 15.9–30.7%, p<0.001). The placebo response of patients predicted to be high placebo responders (placebo response score higher than the mean) was substantially higher than the placebo response of patients predicted to be placebo nonresponders (placebo response score lower than the mean) (Figure 3A). Using this placebo response score as a covariate resulted in a decrease in data variability that translated into a 37.2-percent improvement in the precision of the treatment effect estimation, demonstrating the tool’s effectiveness in increasing clinical trial assay sensitivity (Figure 3B).46
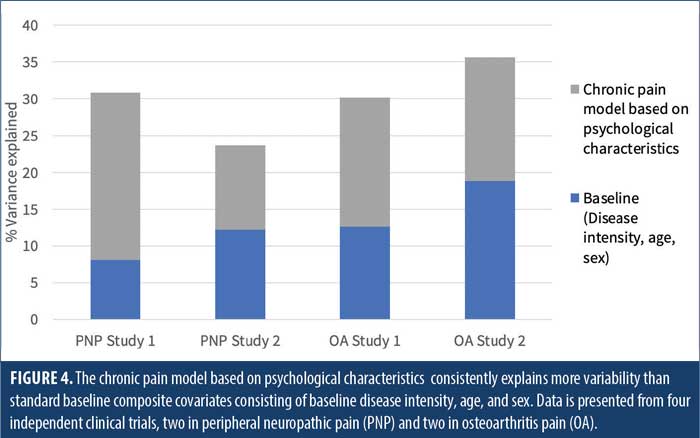
A similar approach was taken to build a predictive model of placebo response in Parkinson’s disease in a cohort of 94 patients. This approach predicted placebo response for multiple endpoints, including Unified Parkinson’s Disease Rating Scale III (UPDRS III) (Pearson’s correlation=66.0%, R2=43.6%, population R2=33.2%, p<0.001).47 Importantly, this covariate approach explains 2- to 3-fold more variability in placebo response than standard factors, including baseline disease intensity, age, and sex (Figure 4) across multiple trials in PNP and OA pain, demonstrating the critical importance of including personality traits in predictive modeling of placebo response.
Regulatory Considerations
With the increasing use of AI and ML in clinical drug development, regulatory agencies are engaged in reflection papers and discussions with stakeholders, including the pharmaceutical industry, academia, scientific consortia, and patient advocacy groups.48–55 Recently, regulatory agencies have issued proposed frameworks for using AI/ML algorithms in clinical trials56 and requested stakeholder feedback.57,58 These interactions highlight the importance of better defining the framework and further characterizing the issues surrounding the development and use of AI/ML algorithms.
While regulators are aware of some of the advantages these AI technologies might offer in optimizing drug development, it is essential that they establish policies and develop guidance on this topic. For example, the FDA’s Center for Devices and Radiological Health and the European Union (EU) have respectively published their action plans for the use of AI/ML-based software as a medical device57 and a fact sheet on Medical Device Software (MDSW),59 as well as the 2021 Coordinated Plan on AI, which is the next step in creating EU-based global leadership in trustworthy AI.60
In the US, the FDA’s Center for Drug Evaluation Research Critical Path Innovation Meetings61 remains a forum for stakeholders to share their approaches and seek guidance on the best path forward with technologies enhancing clinical drug development. When these AI methods are intended to be applied to specific drug development programs, sponsors are recommended to seek the opinion of the relevant FDA review division at an early stage of planning under their investigational new drug (IND) application. This is particularly important, for instance, if a sponsor intends to use AI/ML in the enrichment of the proposed study population (e.g., selection of placebo nonresponders in a CNS indication). Discussions should include the appropriateness of the proposed patient selection criteria, any detection of drug effect difference in that selected/enriched population (as compared to that in the unselected population), and how to reflect such trial results in product labeling.62 In Europe, the European Medicines Agency (EMA) has several venues in which scientists are encouraged to present new technologies enhancing clinical development, in particular the Innovation Task Force63 and the Qualification Advice process.64
With respect specifically to placebo response prediction using AI/ML, agencies are fully aware of the negative impact of the placebo response on clinical development efficiency. In the context of Digital Health Technologies (DHT), the focus of the EMA qualification process (which might also be applicable to the FDA expectations) is to assess whether the measure taken with the technology is fit for its intended clinical use in the regulatory context of medicinal products development/evaluation. Applicants are expected to elaborate on the clinically meaningful relevance of the concept of interest and on the potential benefit of using new technology measures over other existing methods.
To avoid any bias in the data interpretation and to preserve accuracy, usability, equity, and trust, regulatory agencies will expect full transparency and disclosure of the modeling (e.g., feature selection methodologies, proven predictivity, and reproducibility of the models in independent clinical trials). When applicable, prespecification of the model must be documented before conducting the analysis. For example, methods applied to validate and test the algorithms and their accuracy, reliability, and sensitivity to change will be required. Any exclusion of patients from the population eligible for RCTs might be reflected in the labeling, unless extrapolation is accepted. Furthermore, patient exclusion based on increased likelihood of placebo response might be ethically or scientifically unacceptable, for the reasons mentioned above, and so such patient exclusion strategies would have to be justified and, ideally, decided upon after early interaction with regulators.
Conclusion
The placebo response has challenged drug development for decades, making it difficult to distinguish true treatment efficacy from placebo response. Techniques such as training sites to avoid inflating patient expectation in patient interactions and identifying placebo responders through placebo run-in periods have, in some cases, provided modest improvement in placebo response rates; yet, the placebo response is continuing to increase in many CNS indications. Even if these techniques have marginally succeeded to limit the negative impact of the placebo response, they have not significantly improved characterization of drug effect size.
The emergence of advanced data analytic techniques (AI) creates a significant opportunity to influence drug development and clinical research. These techniques are not only beneficial, but they might also be necessary to address a phenomenon as complex as placebo response. While the drug development community is just beginning to capitalize on the power of AI to better understand and limit the negative consequences of the placebo response, this field is truly in its infancy. Even so, the analysis and examples presented here demonstrate the real potential of an AI-based approach, ranging from generating meaningful associations in depression and bipolar disorder to developing predictive algorithms in schizophrenia, pain, and Parkinson’s disease. This area will continue to evolve over time, with further efforts needed for this approach to reach its full potential.
All stakeholders in drug development, including pharmaceutical and biotechnology companies, contract research organizations (CROs), technology developers, patients, regulators, and payers, can and should play active roles in furthering the use of AI to better understand and ultimately predict placebo response. Collaboration and cooperation between sponsor companies to share and pool clinical trial data would be significantly enabling, acknowledging that standards for data organization and quality are critical for this to be effective. Technology developers can work to accelerate acceptance of these approaches by providing more use cases and demonstration of positive impact. Regulators can facilitate the adoption of such approaches by providing clear paths for their evaluation and ensuring their appropriate, valid, and ethical use.
In the post-COVID-19 era of drug development, we predict that a new era of innovation will be born, with AI-based approaches increasing in utility and acceptance. Industry trends focusing on innovation are already emerging. The advent of digital endpoints is providing increased data resolution, but also requiring new tools to manage massive datasets. Decentralized, virtual, and hybrid trials are increasing across the industry, but also raising questions of how this new trial context will impact placebo response rates. The use of AI to address these and other issues has the potential to positively impact trial success rates, cost, timelines, and the translatability of clinical trial data to larger patient populations, with benefits apparent to all.
Acknowledgments
The authors acknowledge Venkatesha Murthy and Elizabeth Hanson from Takeda Pharmaceuticals for contributions to the unpublished data in the “Associating placebo response with attitudinal variables in bipolar disorder” section. The authors acknowledge Alvaro Pereira, Samuel Branders, Chantal Gossuin, and Frederic Clermont from Tools4Patient and Jamie Dananberg from Unity Biotechnology for contributions to the unpublished data in the “Prediction of patient placebo responsiveness using psychological traits in multiple diseases” section. The authors wish to thank Mary Bea Harding, Linda Hutchinson, and D’Laine Bennet for their help with organizing the International Society for CNS Clinical Trials and Methodology (ISCTM) AI/ML in placebo response working group meetings and assisting with administrative tasks.
Disclaimer
This position paper is the work product of the ISCTM “Use of AI/ML in Placebo Response” working group that began at the 16th Annual ISCTM Scientific Meeting in February 2020 in Washington, DC. This working group is part of a larger working group titled “Artificial Intelligence (AI) and Machine Learning (ML) for CNS Clinical Trials,” chaired by Adam Butler and Larry Alphs.
This article reflects the views of the authors and should not be understood or quoted as being made on behalf or reflecting the position of the agencies or organizations with which the authors are affiliated.
The views expressed in this article are the personal views of the author(s) and should not be understood or quoted as being made on behalf of or reflecting the position of the regulatory agency/agencies or organizations with which the author(s) is/are employed/affiliated.
Dr. Khin contributed to this work while she was employed with the FDA. Dr. Khin is now employed by Neurocrine Biosciences, Inc. in San Diego, California.
References
- Bridge JA, Birmaher B, Iyengar S, et al. Placebo response in randomized controlled trials of antidepressants for pediatric major depressive disorder. Am J Psychiatry. 2009;166(1):42–49.
- Kasper S, Dold M. Factors contributing to the increasing placebo response in antidepressant trials. World Psychiatry. 2015;14(3):304–306.
- Tuttle AH, Tohyama S, Ramsay T, et al. Increasing placebo responses over time in US clinical trials of neuropathic pain. Pain. 2015;156(12):2616–2626.
- Vieta E, Cruz N. Increasing rates of placebo response over time in mania studies. Psychiatrist.com. https://www.psychiatrist.com/jcp/assessment/research-methods-statistics/increasing-rates-placebo-response-mania-studies/. Accessed 23 May 2021.
- Gopalakrishnan M, Zhu H, Farchione TR, et al. The trend of increasing placebo response and decreasing treatment effect in schizophrenia trials continues: an update from the US Food and Drug Administration. J Clin Psychiatry. 2020;81(2):19r12960.
- Khin NA, Chen YF, Yang Y, et al. Exploratory analyses of efficacy data from schizophrenia trials in support of new drug applications submitted to the US Food and Drug Administration. J Clin Psychiatry. 2012;73(6):856–864.
- Khin NA, Chen YF, Yang Y, et al. Exploratory analyses of efficacy data from major depressive disorder trials submitted to the US Food and Drug Administration in support of new drug applications. J Clin Psychiatry. 2011;72(4):464–472.
- Häuser W, Bartram-Wunn E, Bartram C, et al. Systematic review: placebo response in drug trials of fibromyalgia syndrome and painful peripheral diabetic neuropathy–magnitude and patient-related predictors. Pain. 2011;152(8):1709–1717.
- Rief W, Nestoriuc Y, Weiss S, et al. Meta-analysis of the placebo response in antidepressant trials. J Affect Disord. 2009;118(1–3):1–8.
- Rutherford BR, Roose SP. A model of placebo response in antidepressant clinical trials. Am J Psychiatry. 2013;170(7):723–733.
- Benedetti F, Carlino E, Piedimonte A. Increasing uncertainty in CNS clinical trials: the role of placebo, nocebo, and Hawthorne effects. Lancet Neurol. 2016;15(7):736–747.
- Wager TD, Atlas LY. The neuroscience of placebo effects: connecting context, learning and health. Nat Rev Neurosci. 2015;16(7):403–418.
- Kirsch I. Response expectancy and the placebo effect. Int Rev Neurobiol. 2018;138:81–93.
- Papakostas GI, Fava M. Does the probability of receiving placebo influence clinical trial outcome? A meta-regression of double-blind, randomized clinical trials in MDD. Eur Neuropsychopharmacol. 2009;19(1):34–40.
- Darragh M, Booth RJ, Consedine NS. Who responds to placebos? Considering the “placebo personality” via a transactional model. Psychol Health Med. 2015;20(3):287–295.
- Weimer K, Colloca L, Enck P. Placebo effects in psychiatry: mediators and moderators. Lancet Psychiatry. 2015;2(3):246–257.
- Fava M, Evins AE, Dorer DJ, Schoenfeld DA. The problem of the placebo response in clinical trials for psychiatric disorders: culprits, possible remedies, and a novel study design approach. Psychother Psychosom. 2003;72(3):115–127.
- van Seventer R, Bach FW, Toth CC, et al. Pregabalin in the treatment of post-traumatic peripheral neuropathic pain: a randomized double-blind trial. Eur J Neurol. 2010;17(8):1082–1089.
- Hulshof TA, Zuidema SU, Gispen-de Wied CC, Luijendijk HJ. Run-in periods and clinical outcomes of antipsychotics in dementia: a meta-epidemiological study of placebo-controlled trials. Pharmacoepidemiol Drug Saf. 2020;29(2):125–133.
- Trivedi MH, Rush J. Does a placebo run-in or a placebo treatment cell affect the efficacy of antidepressant medications? Neuropsychopharmacol. 1994;11(1):33–43.
- Lee S, Walker JR, Jakul L, Sexton K. Does elimination of placebo responders in a placebo run-in increase the treatment effect in randomized clinical trials? A meta-analytic evaluation. Depress Anxiety. 2004;19(1):10–19.
- United States Food and Drug Administration. FDA briefing document Psychopharmacologic Drugs Advisory Committee (PDAC) and Drug Safety and Risk Management (DSaRM) advisory committee meeting. 2018. https://www.fda.gov/media/121359/download. Accessed 21 Jan 2022.
- Zilcha-Mano S, Roose SP, Brown PJ, Rutherford BR. A machine learning approach to identifying placebo responders in late-life depression trials. Am J Geriatr Psychiatry. 2018;26(6):669–677.
- Trivedi MH, South C, Jha MK, et al. A novel strategy to identify placebo responders: prediction index of clinical and biological markers in the EMBARC trial. 2018. www.karger.com/pps. Accessed 2021 Feb 9.
- Lorenzo-Luaces L, Rodriguez-Quintana N, Riley TN, Weisz JR. A placebo prognostic index (PI) as a moderator of outcomes in the treatment of adolescent depression: could it inform risk-stratification in treatment with cognitive-behavioral therapy, fluoxetine, or their combination? Psychother Res. 2021;31(1):5–18.
- Zilcha-Mano S, Roose SP, Brown PJ, Rutherford BR. Abrupt symptom improvements in antidepressant clinical trials: transient placebo effects or therapeutic reality? J Clin Psychiatry. 2019;80(1):E1–E6.
- Zilcha-Mano S, Brown PJ, Roose SP, et al. Optimizing patient expectancy in the pharmacologic treatment of major depressive disorder. Psychol Med. 2019;49(14):2414–2420.
- Sikora M, Heffernan J, Avery ET, et al. Salience network functional connectivity predicts placebo effects in major depression. Biol Psychiatry Cogn Neurosci Neuroimaging. 2016;1(1):68–76.
- Tu Y, Ortiz A, Gollub RL, et al. Multivariate resting-state functional connectivity predicts responses to real and sham acupuncture treatment in chronic low back pain. Neuroimage Clin. 2019;23:101885.
- Anderson AE, Keefe R, Fu DJ, et al. Trials, errors, and placebo prediction: tradeoffs between effect size and sample size in machine learning models to mitigate the placebo response. ISCTM: The International Society for CNS Clinical Trials and Methodology 15th Annual Scientific Meeting. 2019. https://isctm.org/public_access/Feb2019/Posters/Anderson-poster.pdf. Accessed 10 Mar 2022.
- Feltner D, Hill C, Lenderking W, et al. Development of a patient-reported assessment to identify placebo responders in a generalized anxiety disorder trial. J Psychiatr Res. 2009;43(15):1224–1230.
- Qorri B, Tsay M, Agrawal A, et al. Using machine intelligence to uncover Alzheimer’s disease progression heterogeneity. Explor Med. 2020;1(6):377–395.
- Geraci J, Wong B, Ziauddin J, et al. Classical and quantum machine learning applied to predicting placebo response for clinical trials in bipolar disorder: recent results. ISCTM: The International Society for CNS Clinical Trials and Methodology 2018 autumn conference. 2018. https://isctm.org/public_access/Autumn2018/PDFs/Geraci-Poster.pdf. Accessed 21 Jan 2022.
- Peiris N, Blasini M, Wright T, Colloca L. The placebo phenomenon: a narrow focus on psychological models. Perspect Biol Med. 2018;61(3):388–400.
- Vachon-Presseau E, Berger SE, Abdullah TB, et al. Brain and psychological determinants of placebo pill response in chronic pain patients. Nat Commun. 2018 ;9(1):1–15.
- Corsi N, Colloca L. Placebo and nocebo effects: the advantage of measuring expectations and psychological factors. Front Psychol. 2017;8:308.
- Petersen GL, Finnerup NB, Grosen K, et al. Expectations and positive emotional feelings accompany reductions in ongoing and evoked neuropathic pain following placebo interventions. Pain. 2014;155(12):2687–2698
- Darragh M, Chang JW-H, Booth RJ, Consedine NS. The placebo effect in inflammatory skin reactions: the influence of verbal suggestion on itch and weal size. J Psychosom Res. 2015;78(5):489–494.
- Bartels DJP, van Laarhoven AIM, van de Kerkhof PCM, Evers AWM. Placebo and nocebo effects on itch: effects, mechanisms, and predictors. Eur J Pain. 2016;20(1):8–13.
- Yu R, Gollub RL, Vangel M, et al. Placebo analgesia and reward processing: Integrating genetics, personality, and intrinsic brain activity. Hum Brain Mapp. 2014;35(9):4583–4593.
- Peciña M, Azhar H, Love TM, et al. Personality trait predictors of placebo analgesia and neurobiological correlates. Neuropsychopharmacol. 2013;38(4):639–646.
- Geers AL, Wellman JA, Fowler SL, et al. Dispositional optimism predicts placebo analgesia. J Pain. 2010;11(11):1165–1171.
- United States Food and Drug Administration. Adjusting for covariates in randomized clinical trials for drugs and biological products. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/adjusting-covariates-randomized-clinical-trials-drugs-and-biological-products. Accessed 22 Jul 2021.
- Committee for Medicinal Products for Human Use. Committee for Medicinal Products for Human Use (CHMP) guideline on adjustment for baseline covariates in clinical trials. 2015. www.ema.europa.eu/contact. Accessed 8 Mar 2020.
- Branders S, Pereira A, Smith E, Demolle D. Modeling of peripheral neuropathic pain and osteoarthritis placebo response: working towards a unique model of the placebo response in chronic pain? ISCTM: The International Society for CNS Clinical Trials and Methodology 16th Annual Scientific Meeting. 2020. https://isctm.org/public_access/Feb2020/Posters/Branders-Poster.pdf. Accessed 10 Mar 2022.
- Branders S, Dananberg J, Clermont F, et al. Predicting the placebo response in OA to improve the precision of the treatment effect estimation. Osteoarthritis Cartilage. 2021;29(Suppl 2):S18–S19.
- Branders S, Rascol O, Garraux G, et al. Modeling of the placebo response in Parkinson’s disease. In: Proceedings of the International Parkinson and Movement Disorders Society MDS Virtual Congress; 2021:369.
- Mantua V, Arango C, Balabanov P, Butlen-Ducuing F. Digital health technologies in clinical trials for central nervous system drugs: an EU regulatory perspective. Nat Rev Drug Discov. 2021;(20):83–84.
- International Society for CNS Clinical Trials and Methodology. 16th Annual Scientific Meeting. https://isctm.org/meeting/isctm-16th-annual-scientific-meeting/. Accessed 24 May 2021.
- European Commission. White Paper: On artificial intelligence–a European approach to excellence and trust. 2020. https://ec.europa.eu/info/sites/default/files/commission-white-paper-artificial-intelligence-feb2020_en.pdf. Accessed 24 Jan 2022.
- European Medicines Agency. Joint HMA/EMA workshop on artificial intelligence in medicines regulation. 2021. https://www.ema.europa.eu/en/events/joint-hmaema-workshop-artificial-intelligence-medicines-regulation. Accessed 24 May 2021.
- European Medicines Agency. Artificial intelligence in clinical trials–ensuring it is fit for purpose. YouTube. 2 Dec 2020. https://www.youtube.com/watch?v=T92f8O9QIGU. Accessed 24 May 2021.
- United States Food and Drug Administration. Public workshop–evolving role of artificial intelligence in radiological imaging. 2021. https://www.fda.gov/medical-devices/workshops-conferences-medical-devices/public-workshop-evolving-role-artificial-intelligence-radiological-imaging-02252020-02262020. Accessed 24 May 2021.
- European Commission. Expert group on AI: shaping Europe’s digital future. Updated 27 Sept 2021. https://digital-strategy.ec.europa.eu/en/policies/expert-group-ai. Accessed 24 Jan 2022.
- United States Food and Drug Administration. Statement from FDA Commissioner Scott Gottlieb, MD on steps toward a new, tailored review framework for artificial intelligence-based medical devices. 2 Apr 2019. https://www.fda.gov/news-events/press-announcements/statement-fda-commissioner-scott-gottlieb-md-steps-toward-new-tailored-review-framework-artificial. Accessed 24 May 2021.
- United States Food and Drug Administration. Artificial intelligence and machine learning in software as a medical device. 2021. https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-software-medical-device. Accessed 24 May 2021.
- Danish Medicines Agency. Suggested criteria for using AI/ML algorithms in GxP. 8 Mar 2021. https://laegemiddelstyrelsen.dk/en/licensing/supervision-and-inspection/inspection-of-authorised-pharmaceutical-companies/using-aiml-algorithms-in-gxp/. Accessed 24 May 2021.
- United States Food and Drug Administration. FDA releases artificial intelligence/machine learning action plan. 12 Jan 2021. https://www.fda.gov/news-events/press-announcements/fda-releases-artificial-intelligencemachine-learning-action-plan. Accessed 24 May 2021.
- European Commission. Is your software a medical device? https://ec.europa.eu/health/sites/default/files/md_sector/docs/md_mdcg_2021_mdsw_en.pdf. Accessed 24 May 2021.
- European Commission. Coordinated plan on artificial intelligence 2021 review. https://digital-strategy.ec.europa.eu/en/library/coordinated-plan-artificial-intelligence-2021-review. Accessed 24 May 2021.
- United States Food and Drug Administration. Critical path innovation meetings (CPIM). https://www.fda.gov/drugs/new-drugs-fda-cders-new-molecular-entities-and-new-therapeutic-biological-products/critical-path-innovation-meetings-cpim. Accessed 24 May 2021.
- Millis DH. Machine-learning-derived enrichment markers in clinical trials. 20 Feb 2020. https://isctm.org/public_access/Feb2020/Presentation/Millis-Presentation.pdf. Accessed 24 Jan 2022.
- European Medicines Agency. Innovation in medicines. https://www.ema.europa.eu/en/human-regulatory/research-development/innovation-medicines. Accessed 24 May 2021.
- European Medicines Agency. Qualification of novel methodologies for medicine development. https://www.ema.europa.eu/en/human-regulatory/research-development/scientific-advice-protocol-assistance/qualification-novel-methodologies-medicine-development-0. Accessed 24 May 2021.
- Hróbjartsson A, Gøtzsche PC. Placebo interventions for all clinical conditions. Cochrane Database Syst Rev. 2010;2010(1):CD003974.
- Mitsikostas DD, Blease C, Carlino E, et al. European Headache Federation recommendations for placebo and nocebo terminology. J Headache Pain. 2020;21(1):1–7.
- Howick J, Friedemann C, Tsakok M, et al. Are treatments more effective than placebos? A systematic review and meta-analysis. PLoS ONE. 2013;8(5) e62599.





